Abstract
Background: Gene expression profiling (GEP) by microarrays of diffuse large B-cell lymphoma (DLBCL) allows the determination of cell-of-origin that reflects normal B-cell differentiation, which has enabled the categorization of DLBCL into activated B-cell-like (ABC) and germinal center B-cell-like (GCB) subclasses. However, this does not fully embrace the great diversity of B-cell subtypes. We have recently developed a B-cell-associated gene signature (BAGS) classification for DLBCL where tumors are associated to Naïve, Centrocyte, Centroblast, Memory, and Plasmablast phenotypes. BAGS provides independent prognostic information to the ABC/GCB subclasses and the international prognostic index as well as differences in predicted response to drugs used in routine clinical treatment [1,2]. However, acquisition of data from microarray platforms is expensive, labour intensive, and requires cumbersome data analysis. Recently, the NanoString nCounter technology has been introduced as a tool for easy-to-use GEP with high sensitivity and specificity, suitable for clinical purposes.
Aim: To develop a NanoString assay for BAGS classification - the BAGS2Clinic assay, making BAGS attractive in routine clinical settings.
Methods: The first step of the analysis involved selecting a set of candidate genes based on microarray data by combining four clinical cohorts (n = 971) into a training dataset, profiled on Affymetrix HGU-133 plus 2.0 microarrays and BAGS classified according to the golden standard [1]. We chose to use a hierarchical mixture of regularized logistic regression models [3] on GEPs originating from contrasting tumor samples between BAGS subtypes. The procedure was validated by bootstrapping the combined data to assess performance in terms of accuracy, sensitivity, and specificity on the out-of-bag samples.
Secondly, NanoString probes were designed to efficiently capture the transcript variation using gene maps based on the available microarray data. To ensure robust transfer from microarray to NanoString an independent cohort (n = 20) of fresh frozen tissue with previous GEP data from microarray were profiled on the NanoString probeset, to identify genes for which microarray and NanoString measurements were highly correlated. Thirdly, well-correlated genes were used to establish a cross-platform BAGS classifier using multinomial regression [3]. The classifier was trained on the microarray training dataset to establish gene weights.
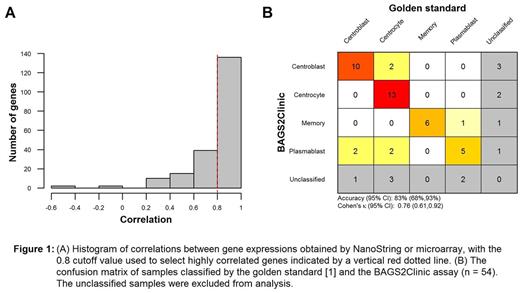
Results: A set of 182 candidate genes were identified by the procedure on the combined cohorts. The accuracy was estimated to be 73% (95% CI 68%-79%) based on the on out-of-bag samples. Of the 182 candidate genes, 136 displayed high correlation between microarray and NanoString measurements (r > 0.8) (Figure 1A). A classifier based on the 136 genes was retrained on the combined data sets and the obtained gene weights are used for the NanoString probes. An accuracy of 83% (95% CI 68%-93%) was obtained when applying the classifier on not previously analysed samples (n = 54) with gene expressions obtained from the NanoString platform (Figure 1B). Furthermore, the sensitivity and specificity of each class versus the rest was good, with ranges from 76%-100% and 89%-100%, respectively.
Conclusion: BAGS2Clinic shows good cross-platform performance. Further studies of BAGS2Clinic are planned on two independent cohorts to validate prognostic and biological differences between BAGS subtypes. This may shed light on the tumor biology as well as differences in resistance to immuno- and chemotherapy, leading to novel treatment strategies of DLBCL patients. In conclusion, this study provides a stepping stone to implement BAGS classification as a routine clinical tool to improve prognosis and treatment guidance for DLBCL patients.
References:
1. Dybkær K, Bøgsted M, Falgreen S, et al. (2015) Diffuse Large B-Cell Lymphoma Classification System That Associates Normal B-Cell Subset Phenotypes With Prognosis. J Clin Oncol , 33(12):1379-1388.
2. Falgreen S, Dybkær K, Young KH, et al. (2015) Predicting response to multidrug regimens in cancer patients using cell line experiments and regularized regression models. BMC Cancer , 2015:15:235.
3. Friedman J, Hastie T, Tibshirani R. (2010) Regularized paths for generalized linear models by coordinate descent. J Stat Soft, 3(1):1-22.
Klapper: Hoffmann La Roche: Other: Grant; Novartis: Other: Grant; Celgene: Other: Grant; Takeda Millenium: Other: Grant. El-Galaly: Roche: Other: Travel funding; Takeda: Other: Travel funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal